3C277.1 e-MERLIN workshop: Part 2 - Imaging
This imaging tutorial follows the calibration tutorial of 3C277.1.
Important: If you have not conducted part 1 (or it went badly), you will need to download the associated scripts and data from the 3C277.1 workshop homepage.
Either way you should have the following files in your current directory before you start
3C277.1_imaging_outline.py- calibration script equivalent of the guide3C277.1_imaging.py- complete script for part 2 in case you get stuck1252+5634.ms- the measurement set (it may need to be extracted if starting at part 2)
How to approach this workshop
The task names are provided in the script and in the content of this tutorial. You need to fill in one or more parameters and values wherever you see '**'. Remember to use the help functionality in CASA, for example, in the case of tclean:
taskhelp # list all tasks
inp(tclean) # possible inputs to task
help(tclean) # help for task
help(par.cell) # help for a particular input (only for some parameters)
This tutorial will be conducted interactively and in script format. You should use the interactive CASA prompt to execute the commands. Once you have completed the task correctly, incorporate the appropriate commands into the 3C277.1_imaging_outline.py script. Alternatively, you can add the correct parameters directly into the script and execute the code step by step (the choice is yours).
Table of Contents
- Initial inspection of 1252+5634.ms (step 1)
- First image of 3C277.1 (step 2)
- Inspect to choose interval for initial phase self-calibration (step 3)
- Derive phase self-calibration solutions (step 4)
- Apply first phase solutions and image again (step 5)
- Phase-self-calibrate again (step 6)
- Apply the solutions and re-image (step 7)
- Choose solution interval for amplitude self-calibration (step 8)
- Amplitude self-cal (step 9)
- Apply amplitude and phase solutions and image again (step 10)
- Reweigh the data according to sensitivity (step 11)
- Try multi-scale clean (step 12)
- Taper the visibilities during imaging (step 13)
- Different imaging weights: Briggs robust (step 14)
- Summary and conclusions
1. Initial inspection of 1252+5634.ms (step 1)
Important: If you have already attempted this tutorial but wish to start from scratch, you can either use clearcal(vis=1252+5634.ms) or, if you believe this has been significantly disrupted, delete it and re-extract the tar file.
Start CASA and list the contents of the measurement set, writing the output to a list file for future reference.
listobs(vis='**',
listfile='**')Make a note of the frequency and the number of channels per spectral window.
You've done this at the end of part 1, but it's always beneficial to practice. Plot the amplitude against the uv distance, averaging all channels per spw (only for correlations RR and LL, both here and for future plotting).
plotms(vis='1252+5634.ms',
xaxis='**',
yaxis='**',
correlation='**',
avgchannel='**',
coloraxis='spw',
overwrite=True)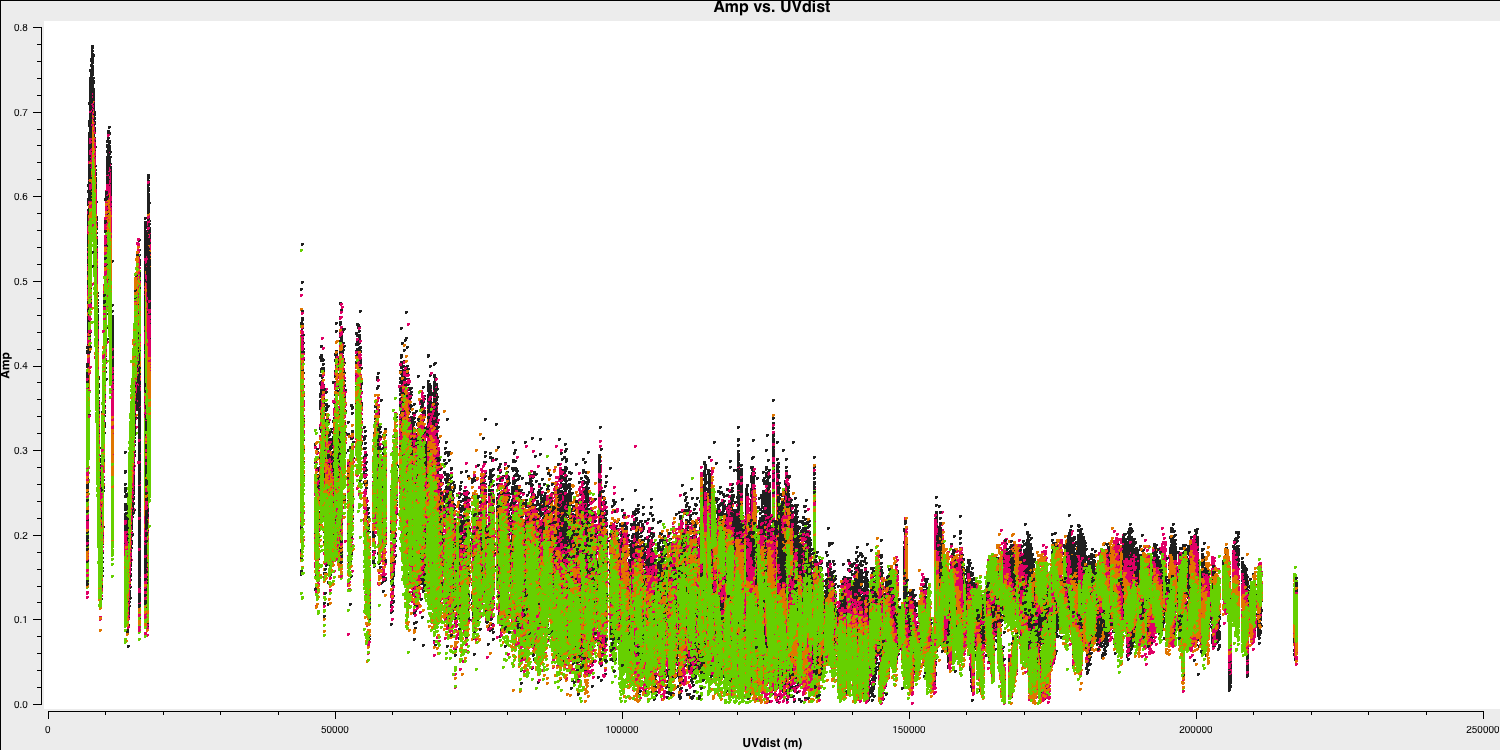
Take note of the $uv$ distance value of the longest baseline, as we will require this for imaging.
Now, in plotms, use the axes tab to select data from the Column drop-down menu and re-plot. The model should default to a flat $1\,\mathrm{Jy}$ if you have not performed any imaging yet.
2. First image of 3C277.1 (step 2)
Now, we need to work out the imaging parameters. We can do this in the CASA prompt through a bit of Python coding:
max_B = *** # max. baseline from uv plot (in metres)
wavelength = *** # approximate observing wavelength (in metres) - listobs gives the frequency.
theta_B= 3600.*degrees(wavelength/max_B) # convert from radians to degrees and then to arcsec
To determine an appropriate cell size, calculate theta_B/5 and round it up to the nearest milliarcsecond. To identify a suitable image size, refer to Ludke et al. 1998 on page 5, and estimate the image size in pixels. Given the sparse $uv$ coverage of e-MERLIN and the resulting extended sidelobes, the optimal image size should be at least twice that to facilitate deconvolution. Since this dataset is relatively small, you can select a power of two that exceeds this size.
Imaging will be conducted using a task called tclean. Important: If you are operating on MacOS, the CASA viewer used for imaging has been deprecated; therefore, we need to use a task called run_iclean instead. If you execute the commands in the CASA prompt or interactive window, you must first import the task with the command from casagui.apps import run_iclean. Likewise, if you exit and re-enter CASA, you should run this command again to make the task available.
Input the derived parameters into the task:
os.system('rm -rf targetp0.*') ## removes any previous imaging run
tclean(vis='1252+5634.ms',
imagename='targetp0',
specmode='mfs',
threshold=0,
cell='**arcsec',
niter=700,
deconvolver='clark',
imsize='***',
interactive=True)
os.system('rm -rf targetp0.*') ## removes any previous imaging run
run_iclean(vis='1252+5634.ms',
imagename='targetp0',
specmode='mfs',
threshold=0,
cell='**arcsec',
niter=700,
deconvolver='clark',
imsize='***')
Set a mask around the clearly brightest areas and clean interactively; mask new areas or enlarge the mask size as necessary. Cease cleaning when no significant flux remains visible.
Examine the image with the CASA task imview (Linux) or CARTA (MacOS). When you click on an image to load, the synthesised beam is displayed, measuring approximately $70\times50\,\mathrm{mas}$ for this image.
Measure the r.m.s. noise and peak brightness. This set of boxes is for a 512-pixel image size, so please change it if you used a different image size.
rms1=imstat(imagename='targetp0.image',
box='10,400,199,499')['rms'][0]
peak1=imstat(imagename='targetp0.image',
box='10,10,499,499')['max'][0]
print(('targetp0.image: peak brightness %7.3f mJy/beam' % (peak1*1e3)))
print(('targetp0.image: rms %7.3f mJy' % (rms1*1e3)))I managed to achieve a peak brightness of $S_p\sim 151.322\,\mathrm{mJy\,beam^{-1}}$ and an rms noise of $\sigma \sim 1.761\,\mathrm{mJy\,beam^{-1}}$. This corresponds to a signal-to-noise ratio ($\mathrm{S/N}$) of around 80. You should find that your S/N is likely similar and falls within the range of 70-120. The rms is highly dependent on the box you use, as the pixel noise distribution is highly non-Gaussian (due to residual calibration errors).
The following command illustrates the use of imview to produce reproducible images (Linux only). This is optional but is shown for reference. You can use imview interactively to decide what values to give the parameters. Note: If you use it interactively, make sure that the black area is a suitable size to just enclose the image, otherwise imview will produce a strange aspect ratio plot.
imview(raster={'file': '1252+5634_1.clean.image',
'colormap':'Rainbow 2',
'scaling': -2,
'range': [-1.*rms1, 40.*rms1],
'colorwedge': True},
zoom=2,
out='1252+5634_1.clean.png')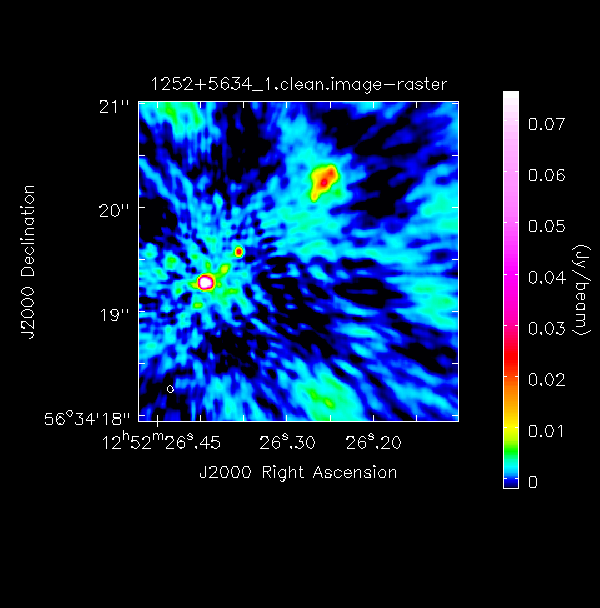
To prepare for self-calibration, we need to inspect the model we will use to derive solutions. The tclean function can perform this automatically if you set the savemodel parameter, but sometimes models are not saved properly. Therefore, we will do this explicitly using the ft task.
ft(vis='***',
model='***',
usescratch=True)
With the $\texttt{MODEL}$ column now filled, use plotms, as in step 1, to plot the model against $uv$ distance.
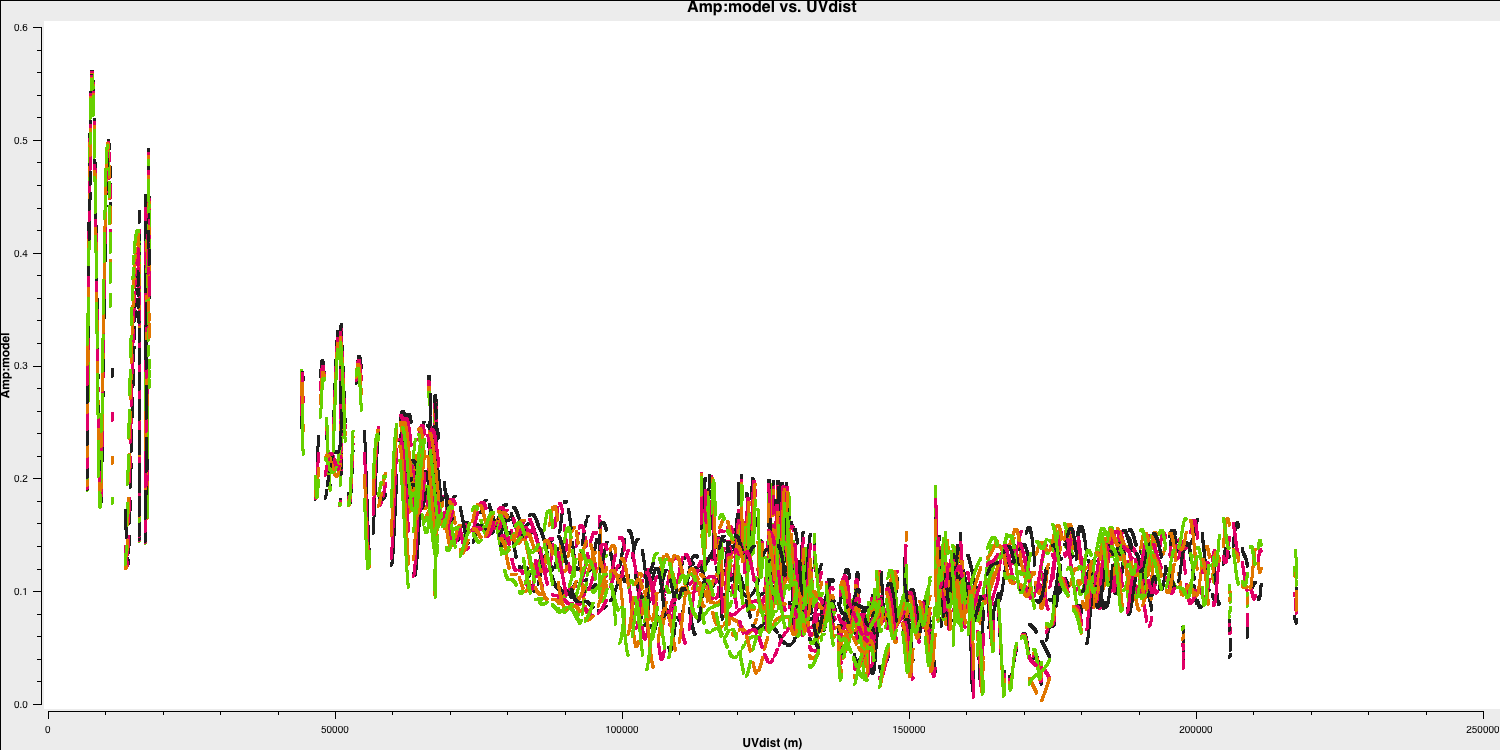
You can also plot data $-$ model which shows that there are still many discrepancies.
3. Inspect to choose interval for initial phase self-calibration (step 3)
It is difficult to distinguish between phase errors and source structure in the visibilities, but it is beneficial to balance between selecting a longer interval to optimise S/N and using an excessively long interval (if possible, the phase changes by less than a few tens of degrees). The phase will change most slowly due to source structure on the shortest baseline, Mk2&Pi, but you should also verify the longest baselines (to Cm) to ensure there is sufficient S/N. Plot the phase against time, averaging all channels per spw (refer to listobs for the number of channels).
plotms(vis='***',
xaxis='***',
yaxis='***',
antenna='***',
correlation='***',
avgchannel='**',
coloraxis='spw',
plotfile='',
timerange='',
overwrite=True)The plot below shows a zoom on several scans. The selected solution interval should ideally fit an integral number of times into each scan. Additionally, it is advisable not to use too short a solution interval for the first model. This is because the model may lack accuracy, so you want to avoid constraining the solutions too tightly.
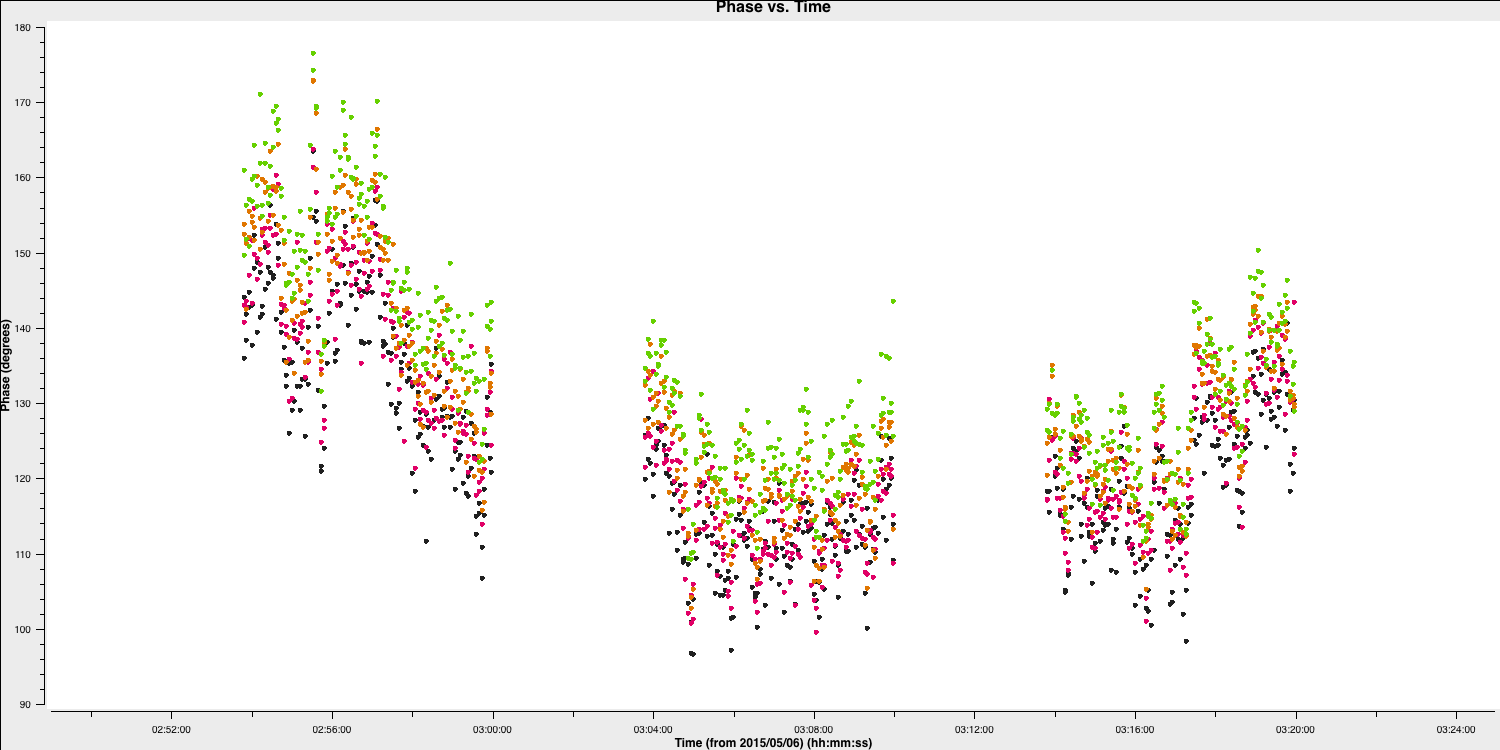
4. Derive phase self-calibration solutions (step 4)
Let's use gaincal to derive solutions based on phase only.
gaincal(vis='1252+5634.ms',
calmode='***', # phase only
caltable='target.p1',
field=target,
solint='***', # about half a scan duration
refant=antref,
minblperant=2, minsnr=2) # These are set to lower values than the defaults which are for arrays with many antennasCheck the terminal and logger; there should ideally be no failed solutions or very few.
Plot the solutions:
plotms(vis='target.p1',
gridcols=2, gridrows=3,
xaxis='time', yaxis='phase',
iteraxis='antenna',
coloraxis='spw',
plotrange=[-1,-1,-180,180],
correlation='L',
showgui=True)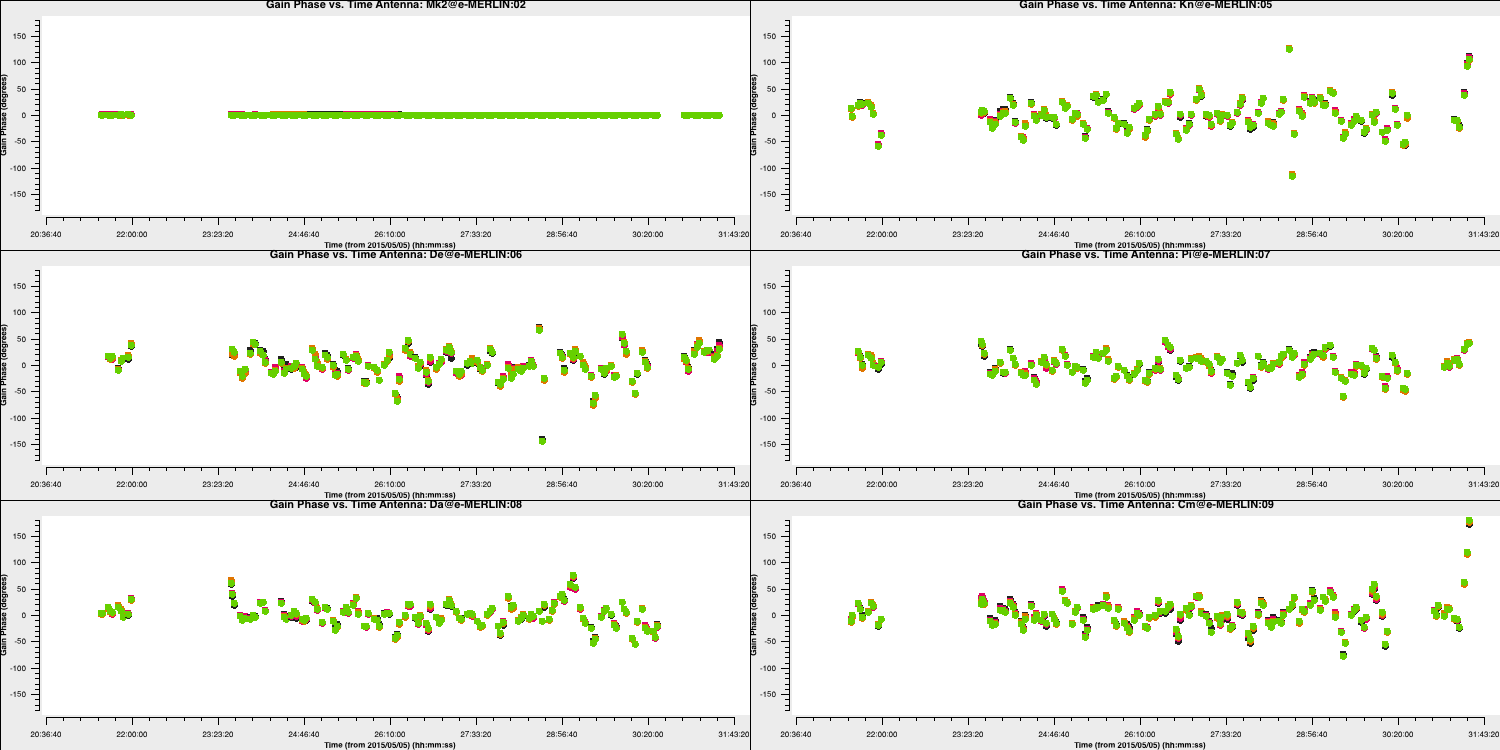
Repeat for the other polarisation. The corrections should remain coherent, as they currently are, and reflect the differences caused by the varying atmospheric paths between the target and phase reference source. However, they may also contain a spurious component if the model is imperfect. The target is bright enough that a shorter solution interval could be used. Therefore, these solutions are applied, the data is re-imaged, and the corrections are refined iteratively.
5. Apply first phase solutions and image again (step 5)
Let's proceed by implementing these solutions for the measurement set. This may take a moment since the $\texttt {CORRECTED}$ data column needs to be generated.
applycal(vis='***',
gaintable='***') # enter the name of the MS and the gaintable
With these corrections applied, let's use tclean as before, but you may be able to perform more iterations since the noise should be lower.
os.system('rm -rf targetp1.*')
tclean(vis='1252+5634.ms',
imagename='targetp1',
specmode='mfs',
threshold=0,
cell='**arcsec',
niter=1500, # stop before this if residuals reach noise
deconvolver='clark',
imsize='***',
interactive=True)os.system('rm -rf targetp1.*')
run_iclean(vis='1252+5634.ms',
imagename='targetp1',
specmode='mfs',
threshold=0,
cell='**arcsec',
niter=1500, # stop before this if residuals reach noise
deconvolver='clark',
imsize='***')
Use imstat to find the r.m.s. again and the viewer/CARTA to inspect the image; you can plot it if you want. Using the same boxes as before, I managed to get $S_p \sim 168.476 \,\mathrm{mJy\,beam^{-1}}$ and $\sigma \sim 0.292\,\mathrm{mJy\,beam^{-1}}$. This is a massive $\mathrm{S/N}$ increase to $\sim 580$.
Important: A key indicator of whether self-calibration is improving your data is that the S/N should increase with each iteration. Do not rely on measuring the rms/peak individually, as the self-calibration model could artificially reduce all amplitudes (peak and noise) without any S/N improvement.
Once more, let's apply the Fourier Transform to this improved model to revert it to the measurement set.
ft(vis='***',
model='***', # remember to use the new model!
usescratch=True)6. Phase-self-calibrate again (step 6)
Now that the model has improved, consider using a shorter solution interval. Heuristics suggest that 30 seconds is generally the minimum worth using, as the atmosphere remains reasonably stable at this frequency over these baseline lengths.
Execute gaincal as before, but decrease the solution interval to 30 seconds and create a new solution table (the new model will be used).
os.system('rm -rf target.p2')
gaincal(vis='1252+5634.ms',
calmode='***',
caltable='target.p2',
field=target,
solint='***',
refant=antref,
gaintable=['***'],
interp=['***'],
minblperant=2, minsnr=2)Plot the solutions:
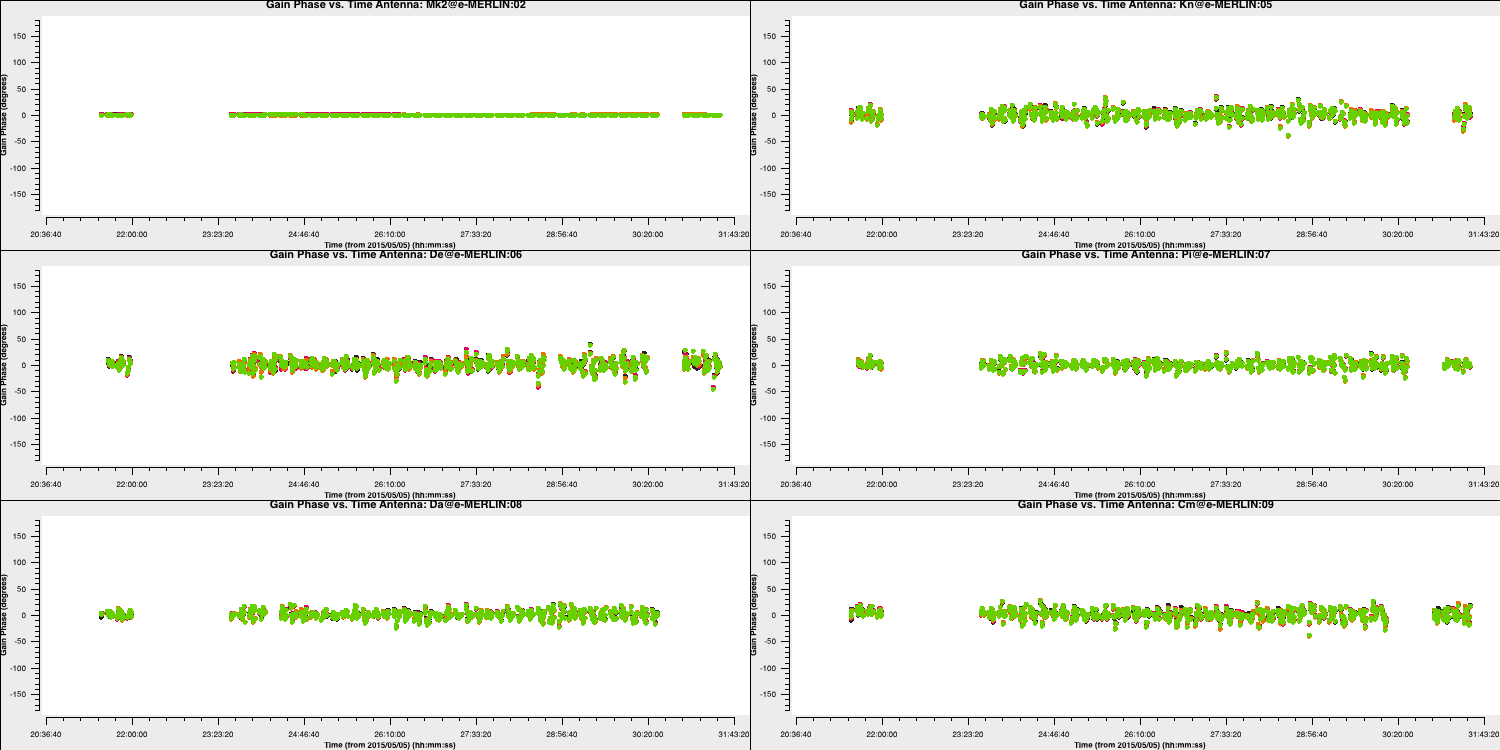
Since we apply the first self-calibration table on the fly, the resulting corrections will be smaller in amplitude than the original corrections. This indicates that the self-calibration is functioning properly, as the phase errors in your data decrease progressively. If you notice that the solutions are noisier, it may signal an issue, such as using a solution interval that is too small.
7. Apply the solutions and re-image (step 7)
Use applycal as before, but making sure we add the new gaintable to the list.
applycal(vis='1252+5634.ms',
gaintable=['***', '***'])
The listobs command indicated that the total bandwidth ranges from 4817 to 5329 MHz. This means that for flux density $S$, the relationship is $S_{4817}/S_{5329} = (4817/5329)^{-1}$, which is approximately 10%. Furthermore, Cotton et al. (2006) finds that the spectral index varies from $-0.1$ in the core to as steep as $-1$. The enhanced signal-to-noise ratio (S/N) provides significantly greater accuracy. Therefore, to properly image and produce an accurate representation for amplitude self-calibration across all spectral windows, multi-term multi-frequency synthesis (MT-MFS) imaging is employed. This technique adjusts the sky model to account for changes in frequency.
Ensure that you clean until as much target flux as possible has been removed from the model - you can interactively increase the number of iterations if desired.
os.system('rm -rf targetp2.*')
tclean(vis='1252+5634.ms',
imagename='targetp2',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=2500, # stop before this if residuals reach noise
imsize='***',
deconvolver='mtmfs',
nterms=2, # Make spectral index image
interactive=True)
os.system('rm -rf targetp2.*')
run_iclean(vis='1252+5634.ms',
imagename='targetp2',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=2500, # stop before this if residuals reach noise
imsize='***',
deconvolver='mtmfs',
nterms=2) # Make spectral index image
Use ls to view the names of the created image files. The name of the image is targetp2.image.tt0.
If we check the image using the same boxes as before, I get $S_p \sim 172.408 \,\mathrm{mJy\,beam^{-1}}$ and $\sigma \sim 0.216\,\mathrm{mJy\,beam^{-1}}$. This is another $\mathrm{S/N}$ increase to $\sim 800$.
Let's incorporate the model we derived into the visibilities. To account for the frequency dependence of the source, we need to include both components of the model (i.e., the tt0 and tt1 models).
ft(vis='1252+5634.ms',
model=['***.model.tt0','***.model.tt1'], # model images from the MTMFS images,
nterms='**', # same as tclean!
usescratch=True)8. Choose solution interval for amplitude self-calibration (step 8)
Next, we aim to correct the amplitude errors. Plot the amplitude against time to select an appropriate solution interval.
plotms(vis='1252+5634.ms',
xaxis='***',
yaxis='***',
ydatacolumn='***',
antenna='Mk2&**',
correlation='**,**',
avgchannel='**',
coloraxis='spw')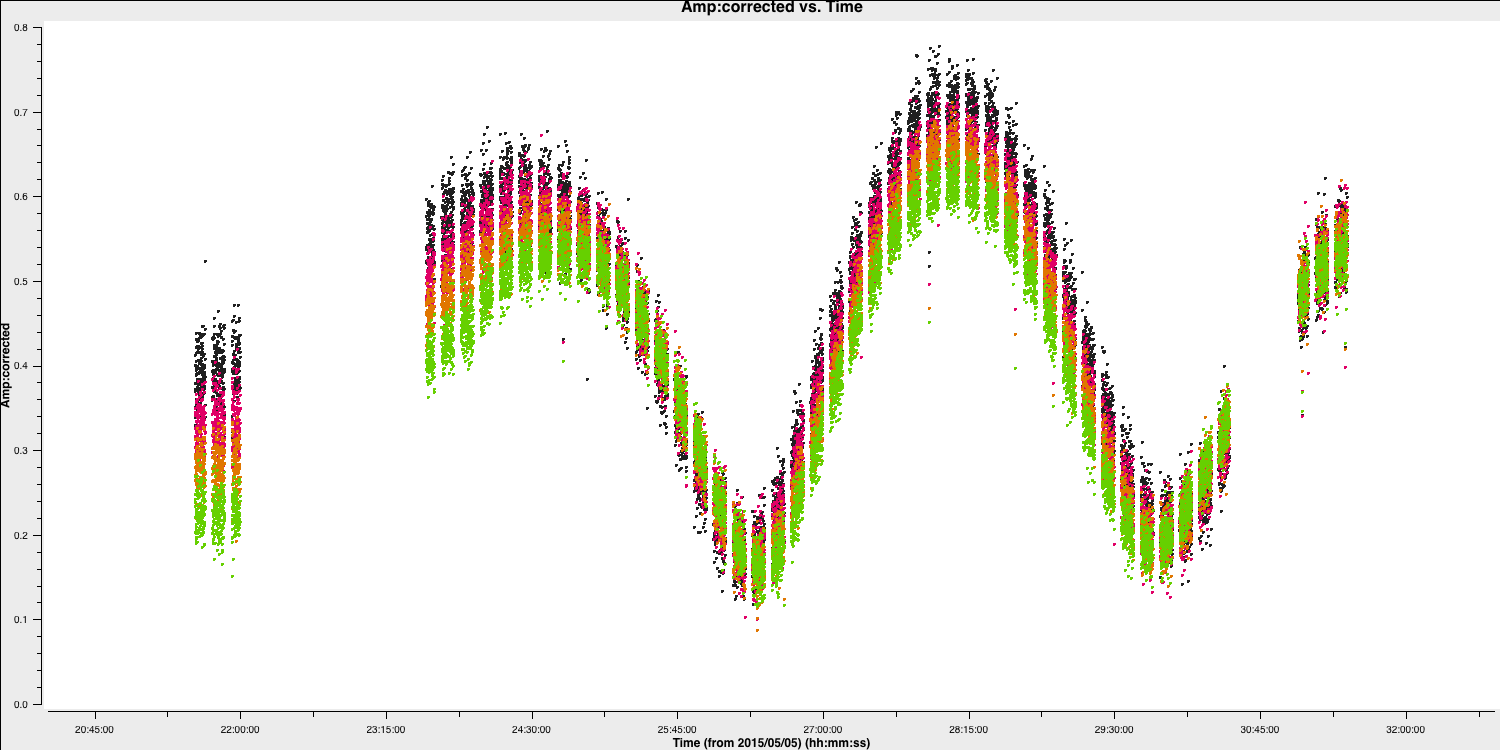
The amplitudes change more slowly than phase errors, and a suitable solution is several times longer than that for phase. Note that the slow, long timescale variations are due to changes in source structure.
9. Amplitude self-cal (step 9)
In gaincal, the previous gaintable with short-interval phase solutions will be applied to allow for a longer solution (averaging) interval for amplitude self-calibration. Set up gaincal to calibrate amplitude and phase, using a longer solution interval and the same reference antenna as before.
os.system('rm -rf target.ap1')
gaincal(vis='1252+5634.ms',
calmode='***', # amp and phase
caltable='target.ap1',
field='1252+5634',
solint='***',
refant='Mk2',
gaintable='***', # the phase table should be applied
minblperant=2,minsnr=2)Plot the solutions, ensuring they are primarily within approximately 20% of unity. A few very low points suggest some bad data (high points). If the solutions consistently exceed unity by a significant margin, then the model is likely missing flux.
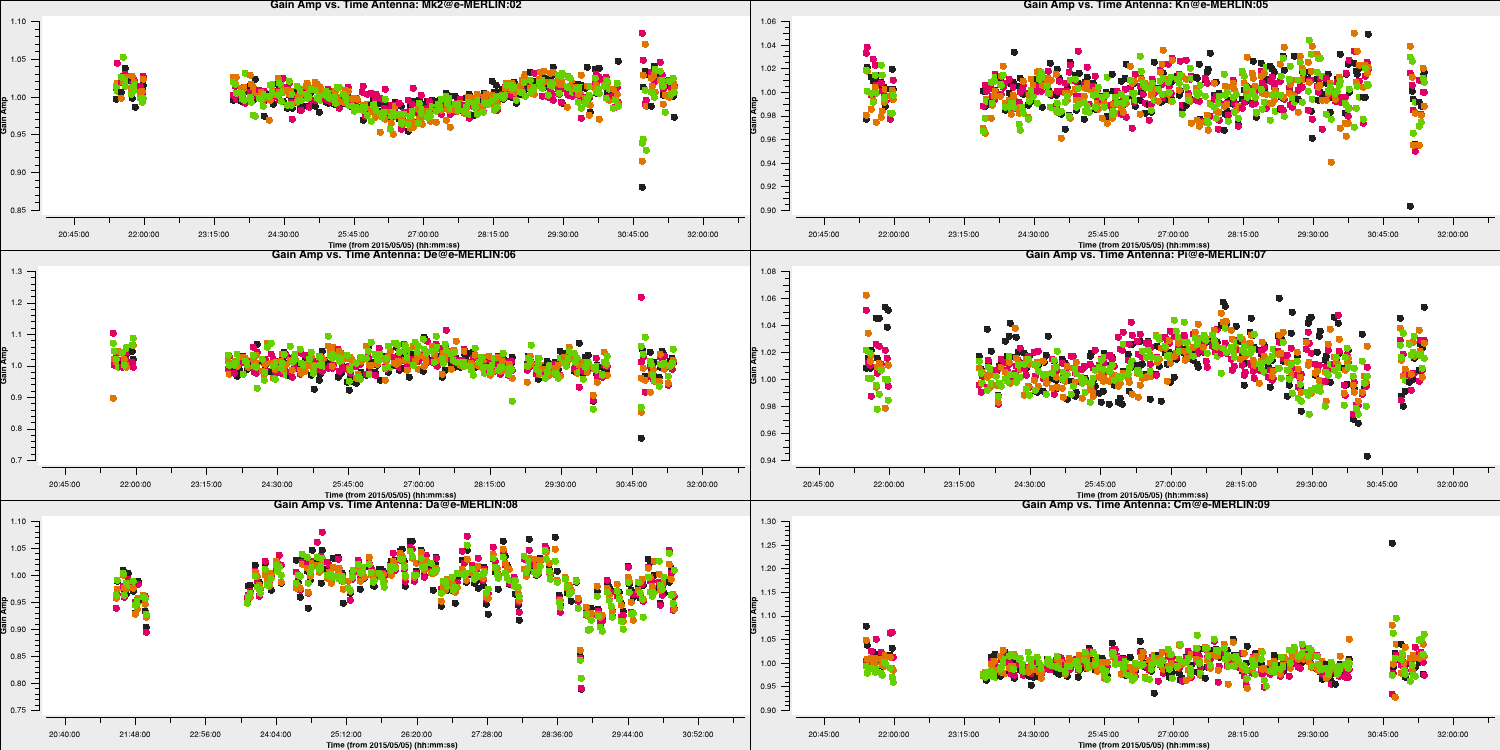
10. Apply amplitude and phase solutions and image again (step 10)
Use applycal to implement the two phase solution tables along with the amplitude and phase table that you have just created.
applycal(vis='1252+5634.ms',
gaintable=['**','**','**'])Re-clean the image as before and inspect it.
os.system('rm -rf targetap1.*')
tclean(vis='1252+5634.ms',
imagename='targetap1',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=3500, # stop before this if residuals reach noise
imsize='***',
deconvolver='mtmfs',
nterms='**', # Make spectral index image
interactive=True)os.system('rm -rf targetap1.*')
run_iclean(vis='1252+5634.ms',
imagename='targetap1',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=3500, # stop before this if residuals reach noise
imsize='***',
deconvolver='mtmfs',
nterms='**') # Make spectral index image
With the same boxes as used before, I get $S_p \sim 171.639 \,\mathrm{mJy\,beam^{-1}}$ and $\sigma \sim 76\,\mathrm{\mu Jy\,beam^{-1}}$. This now corresponds a huge $\mathrm{S/N}\sim 2240$. The key message of self-calibration is to enhance the signal-to-noise ratio (S/N) of your observations by eliminating calibration errors while avoiding constraining your visibilities to the model. Given the number of free parameters, you must adopt an iterative approach to determine the true source structure accurately. It takes practice to discern what is correct and what is not.
11. Reweigh the data according to sensitivity (step 11)
Often, particularly for heterogeneous arrays, the assigned weighting is not optimal for sensitivity. For instance, the Cambridge (Cm) telescope is more sensitive; therefore, to obtain the optimal image, it must be given a larger weight than the other arrays.
To recalculate the optimal weights, we use the task statwt, which calculates the weights based on the scatter in small samples of data. Note that the original data will be overwritten, so you might want to copy the measurement set to a different name or directory first.
There are a lot of options here, but we are just going to use the default settings. You can experiment with different weightings if you'd like.
statwt(vis='***')
In the plot below, we show the weights before (top panel) and after re-weighting (bottom panel) on the baselines to our reference antenna (Mk2). As you can see, the telescopes have similar weights despite Cm being more sensitive. With the use of statwt, the weights now reflect the sensitivities and have also adjusted some errant weights that were low before.
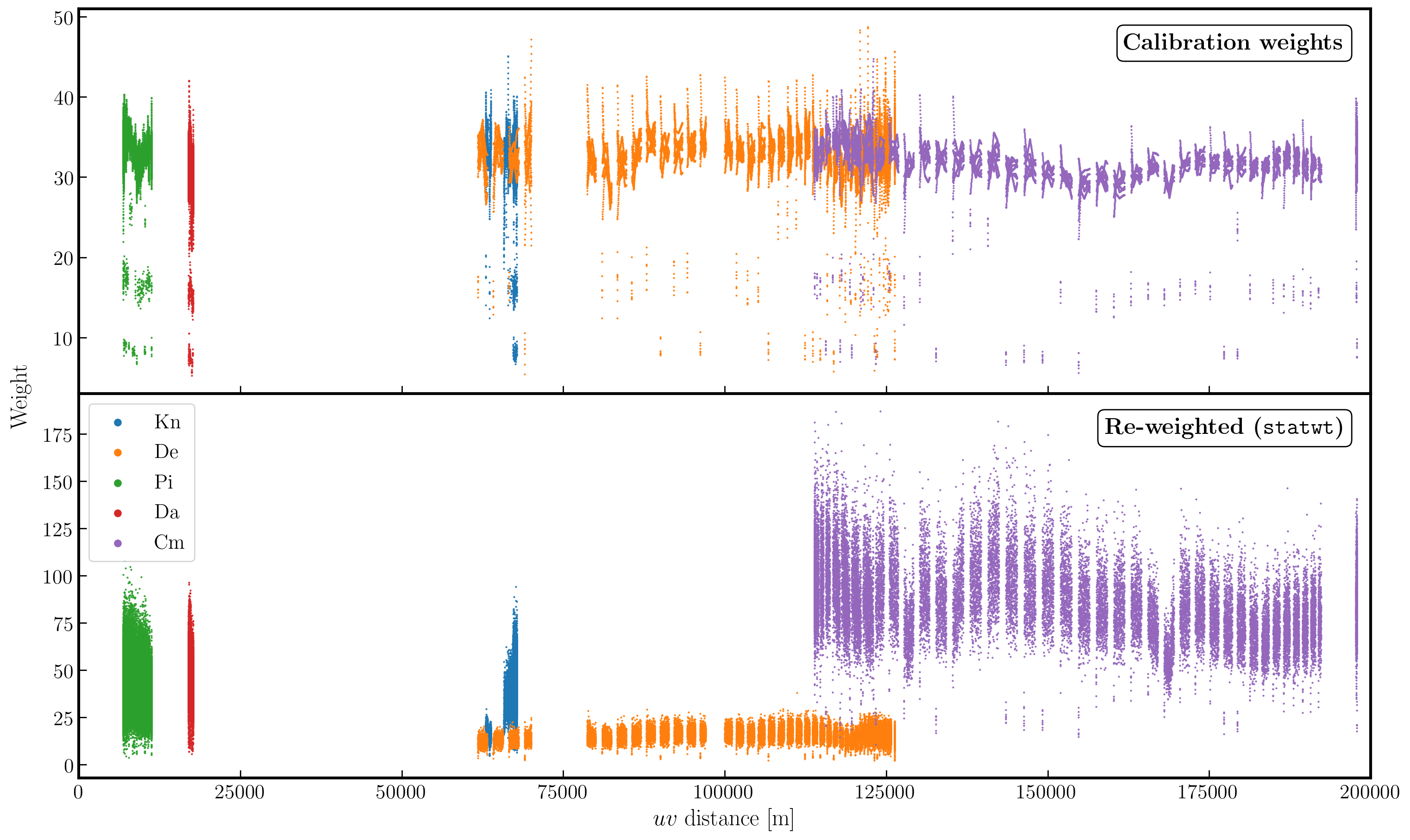
12. Try multi-scale clean (step 12)
Let's image the re-weighted data using multiscale clean, which assumes that the sky can be represented by a collection of Gaussians instead of delta functions. Check the help for tclean, which suggests appropriate values for the multiscale parameter.
os.system('rm -rf targetap1ms.*')
tclean(vis='1252+5634.ms',
imagename='targetap1ms',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter='***', # stop before this if residuals reach noise
imsize='***',
deconvolver='***',
scales=['**','**','**'],
nterms='*',
interactive=True)os.system('rm -rf targetap1ms.*')
run_iclean(vis='1252+5634.ms',
imagename='targetap1ms',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter='***', # stop before this if residuals reach noise
imsize='***',
deconvolver='***',
scales=['**','**','**'],
nterms='*')You will find that the beam is now slightly smaller (I got $65\times53\,\mathrm{mas}$) as the longer baselines have more weight, so the peak brightness is lower (and affects the S/N). I got $S_p \sim 150.381 \,\mathrm{mJy\,beam^{-1}}$ and $\sigma \sim 0.061\,\mathrm{mJy\,beam^{-1}}$.
13. Taper the visibilities during imaging (step 13)
Next, we experiment with changing the relative weights assigned to longer baselines. Applying a taper reduces this, resulting in a larger synthesised beam that is more sensitive to extended emissions. This process only alters the weights during gridding for imaging, with no permanent change to the uv data. However, strong tapering can increase the noise.
Examine the $uv$ distance plot and select an outer taper (half-power of a Gaussian taper applied to the data) that is approximately 2/3 of the longest baseline. The units are in wavelengths, so either calculate that or plot amplitude versus uvwave.
os.system('rm -rf targetap1taper.*')
tclean(vis='1252+5634.ms',
imagename='targetap1taper',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=3500, # stop before this if residuals reach noise
imsize='***',
deconvolver='***',
uvtaper=['***Mlambda'],
scales=['**','**','**'],
nterms='*',
interactive=True)Note that
uvtaper is not currently supported in run_iclean so we do a hard $uv$ distance cut (where we don't select data above a specified $uv$ distance).
os.system('rm -rf targetap1taper.*')
run_iclean(vis='1252+5634.ms',
imagename='targetap1taper',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=3500, # stop before this if residuals reach noise
imsize='***',
deconvolver='***',
uvrange=['<***Mlambda'],
scales=['**','**','**'],
nterms='*')Inspect the image and check the beam size. I got a restoring beam of $74\times61\,\mathrm{mas}$, $S_p \sim 188.140\,\mathrm{mJy\,beam^{-1}}$ and $\sigma \sim 0.105\,\mathrm{mJy\,beam^{-1}}$.
14. Different imaging weights: Briggs robust (step 14)
Now we will experiment with adjusting the relative weights assigned to longer baselines. Default natural weighting assigns zero weight to parts of the uv plane without samples. Briggs weighting interpolates between samples, adding more weight to the interpolated areas for lower values of the 'robust' variable. For an array like e-MERLIN, which has sparser sampling on long baselines, this enhances their contribution, resulting in a smaller synthesised beam. This may highlight the jet/core, possibly at the expense of the extended lobes. Low values of robust increase the noise. You can combine this with tapering or multiscale, but initially, remove these parameters.
os.system('rm -rf targetap1robust.*')
tclean(vis='1252+5634.ms',
imagename='targetap1robust',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=3500, # stop before this if residuals reach noise
imsize='***',
deconvolver='***',
nterms='*',
weighting='***',
robust='**',
interactive=True)os.system('rm -rf targetap1robust.*')
run_iclean(vis='1252+5634.ms',
imagename='targetap1robust',
specmode='mfs',
threshold=0,
cell='***arcsec',
niter=3500, # stop before this if residuals reach noise
imsize='***',
deconvolver='***',
nterms='*',
weighting='***',
robust='**')I got a beam of $56\times40\,\mathrm{mas}$, $S_p \sim 147.872\,\mathrm{mJy\,beam^{-1}}$ and $\sigma \sim 0.191\,\mathrm{mJy\,beam^{-1}}$
15. Summary and conclusions
To summarise, this workshop can be broadly categorised into two main parts: i) self-calibration and ii) imaging.
We started with a simple phase-reference calibrated data set of our target. Here, the phase and amplitude corrections are valid for our nearby phase calibrator, but are only approximately valid for our target. This is because the patch of sky that the telescopes are observing for the target is not the same as that of the phase calibrator.
For our target source, the assumption that it is a simple point source is invalid (as proven by the $uv$ distance plots). This means that in order to calibrate for the gain differences between the phase calibration and target fields, we need to produce a model for calibration. The approximate phase and amplitude solutions are just valid enough for us to derive a first-order model.
Once we generated a model using deconvolution, the Fourier Transform of these models is used for calibration. We begin with phase-only calibration as this requires less signal-to-noise (S/N) for good solutions. With these corrections applied, we can generate a much better model with a dramatically increased S/N. This S/N boost can then be used to calibrate amplitudes and phases. As a result, we create a high dynamic range data set (with a high S/N), where many low-level source structures have emerged. The following table and plot illustrate this vast improvement.
| Peak Brightness | Noise (rms) | S/N | |
|---|---|---|---|
| [$\mathrm{mJy\,beam^{-1}}$] | [$\mathrm{mJy\,beam^{-1}}$] | ||
| Phase referencing | 151.925 | 1.896 | 80.129 |
| Phase self-calibration (cycle 1) | 169.130 | 0.807 | 209.579 |
| Phase self-calibration (cycle 2) | 171.360 | 0.443 | 386.817 |
| Amp+phase self-calibration | 171.717 | 0.174 | 986.879 |
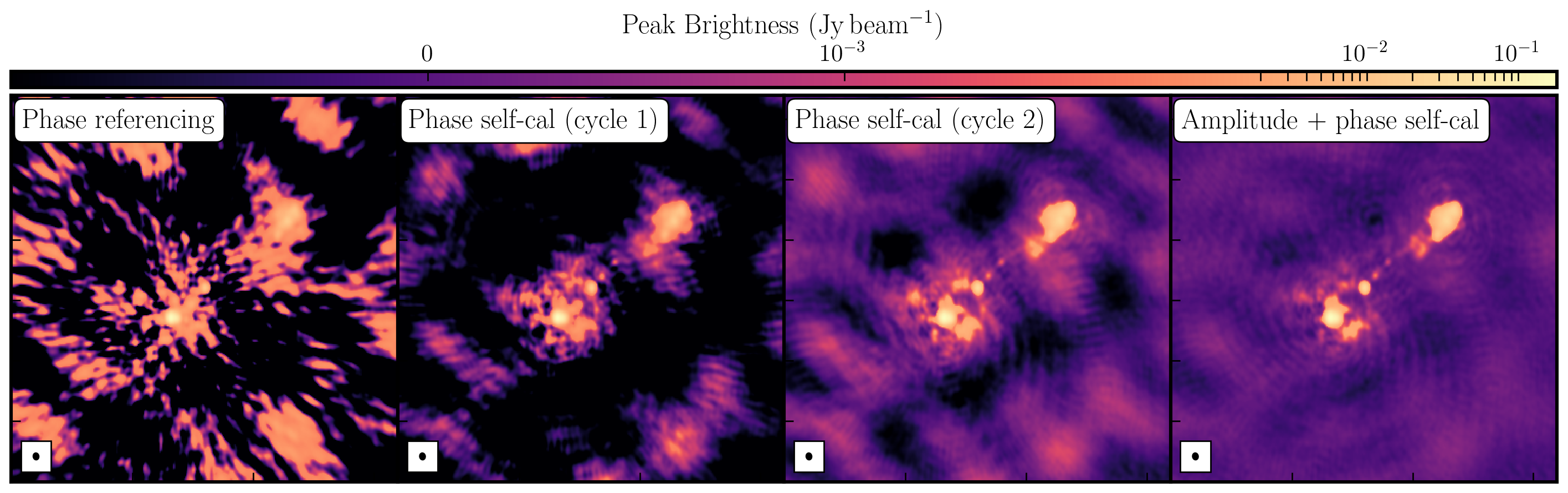
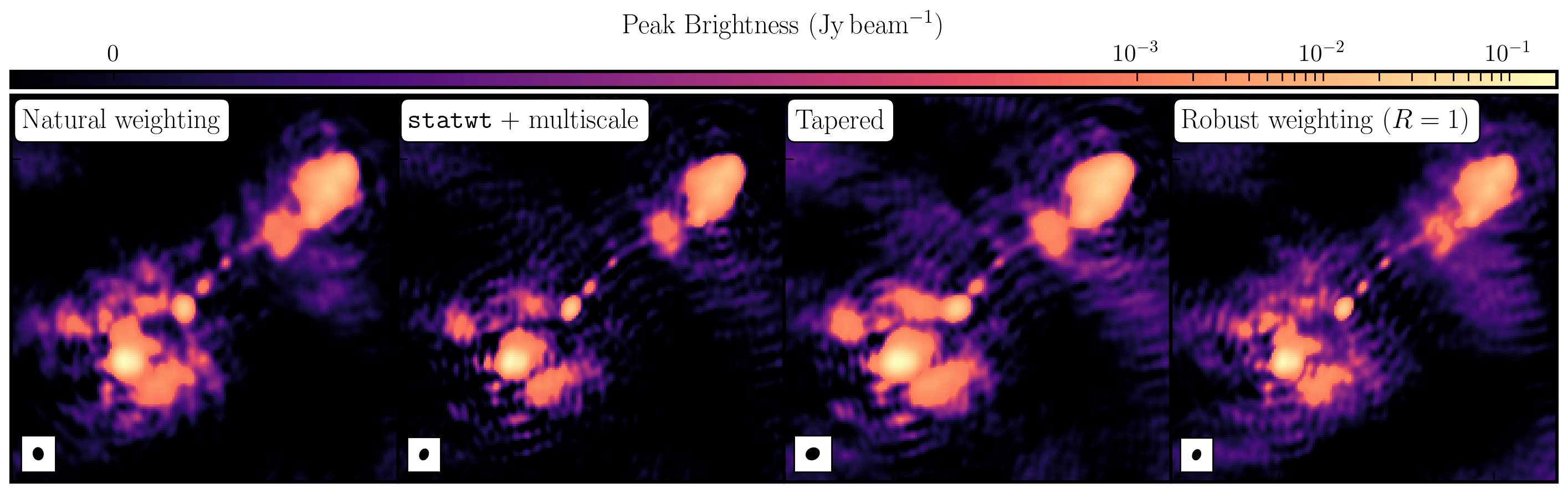
While the first part of this tutorial addressed residual calibration errors related to approximate gain and phase solutions derived from the phase calibrator, the second part explored the different imaging techniques that can be used to enhance features. Initially, we utilized statwt to optimally reweight the data for maximum sensitivity. We find that the Cm telescope is the most sensitive (due to its 32m width) and also includes all the long baselines. Consequently, these long baselines become more prominent when imaging, resulting in a higher resolution image than what we've seen before. As a result, the peak flux decreases, but so does the sensitivity! In addition, we employed multiscale clean to better deconvolve the large, fluffy structures that standard clean can struggle with.
The next step we took was to taper the weights according to the $uv$ distance. The downweighting of the long baselines means that we are more sensitive to larger structures, which results in lower resolution. However, due to the sensitivity inherent in these long baselines, we experience a much larger RMS.
Finally, we introduced robust weighting schemes. These schemes offer a clear method to achieve intermediate resolutions between naturally weighted and uniformly weighted data.
| Peak Brightness | Noise (rms) | S/N | Beam size | |
|---|---|---|---|---|
| [$\mathrm{mJy\,beam^{-1}}$] | [$\mathrm{mJy\,beam^{-1}}$] | [$\mathrm{mas\times mas}$] | ||
| Amp+phase self-calibration | 171.717 | 0.174 | 986.879 | 65 $\times$ 53 |
statwt+multiscale |
151.086 | 0.0760 | 1987.974 | 60 $\times$ 44 |
| Tapering | 188.140 | 0.105 | 1791.810 | 74 $\times$ 61 |
| Robust weighting ($R=1$) | 147.872 | 0.191 | 774.199 | 56 $\times$ 40 |